Predict the effect of BARD1 single nucleotide variants on RNA abundance and cell survival
Challenge: BARD1
Variant data: registered users only
Last updated: 19 October 2025
This challenge is closed. The challenge closed on October 15, 2025.
How to participate in CAGI7? Download data & submit predictions on Synapse
Make sure you understand our Data Use Agreement and Anonymity Policy
Summary
BARD1 (BRCA1-Associated RING Domain 1) forms a heterodimer with BRCA1, which is critical for DNA double-strand break repair and tumor suppression. In this challenge, all possible BARD1 single nucleotide variants were assessed for their effects on RNA abundance and cell survival using Saturation Genome Editing in haploid human cells. Participants are asked to predict two separate function scores for each variant, reflecting experimental measurements of RNA abundance and cellular fitness.
Background
BARD1 is best known for its role in DNA repair and tumor suppression (Irminger-Finger et al, 2016; Tarsounas & Sung, 2020). It contains a RING domain essential for BRCA1 interaction, an ankryin repeat domain, and two tandem BRCT domains (Baer & Ludwig, 2002). Germline mutations in BARD1 are linked to hereditary breast and ovarian cancer. BARD1 assaying reflects the activities of the IGVF Consortium (2024).
Experiment
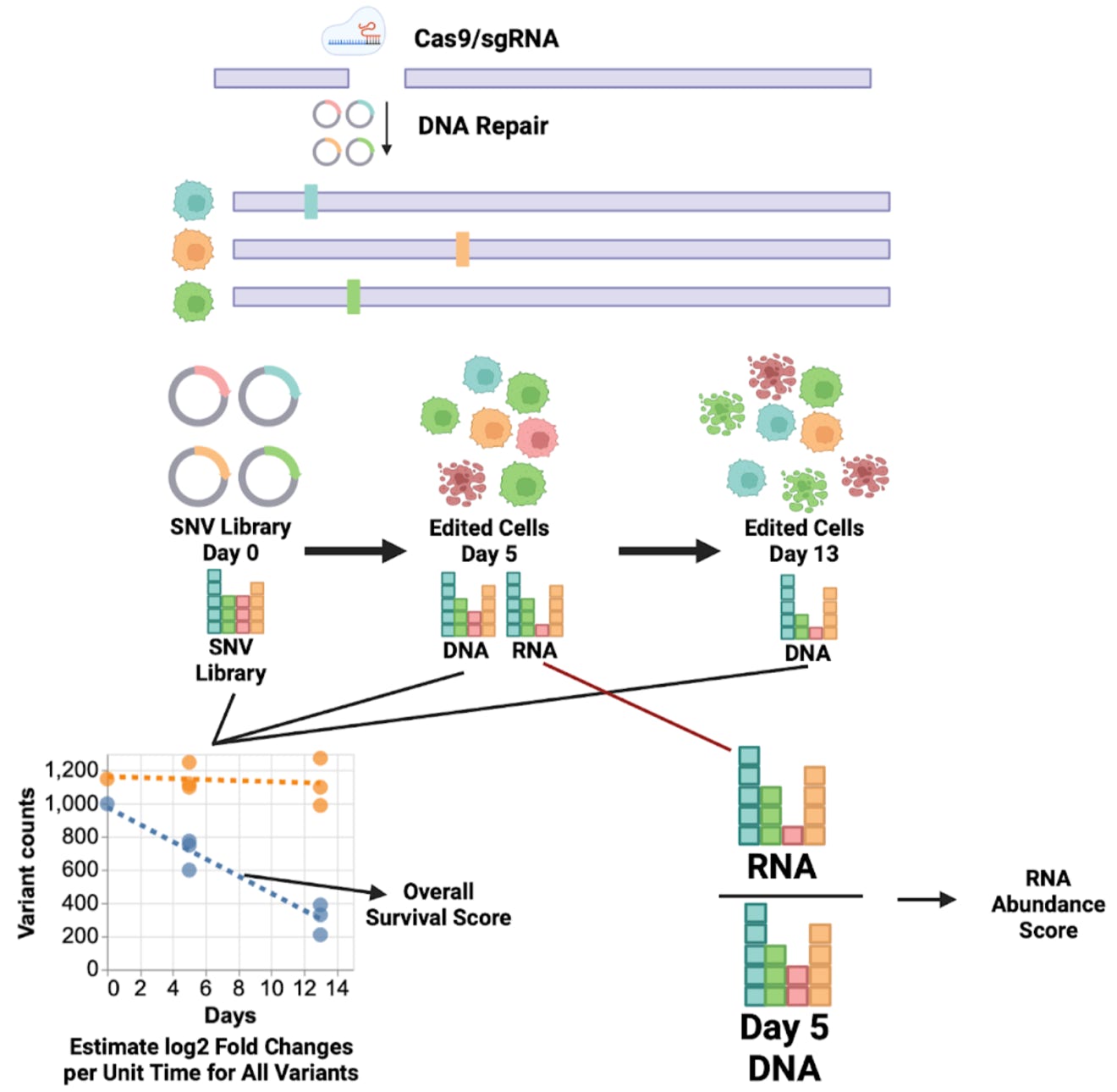
Saturation Genome Editing (SGE) is a cell growth assay enabling systematic interrogation of all possible single nucleotide variants (SNVs) in genes essential for viability of the haploid HAP1 cell line. Variants are introduced via CRISPR/Cas9-mediated homology-directed repair and variant frequencies are quantified through sequencing at two time points (Findlay et al., 2018).
Library design: Saturation genome editing installs all possible variants into a targeted region. All exons, intron/exon boundaries and UTR variants were assessed.
Delivery: Variant libraries and CRISPR/Cas9 gRNAs were chemically transfected into Cas9 expressing HAP1 cells. Transfected cells were selected under puromycin and blasticidin to select for successfully transfected cells and Cas9 expression. Variants were installed in the genome by CRISPR/Cas9-mediated homology-directed repair.
Sampling: The original variant library and successfully transfected cells were sampled 5 and 13 days post-transfection for deep sequencing. gDNA was extracted from both timepoints for sequencing. RNA was also extracted from day 5 samples for targeted RNA sequencing to analyze RNA abundance.
Quantification:
- Overall survival: Variants are identified and counted by sequencing the original variant library (considered day 0) and the edited region at day 5 and day 13 timepoints. Function scores were computed using a negative binomial model to calculate continuous log2 fold changes for each variant across replicates. A score of 0 indicates no change to variant frequency during SGE. Scores <0 indicate decreasing frequency during SGE and possible BARD1 loss-of-function.
- RNA abundance: RNA from day 5 samples was also sequenced. A ratio of day 5 RNA to day 5 DNA frequency was calculated for variants to generate RNA scores. RNA scores <1 have abnormal RNA abundance.
Prediction challenge
Participants must predict both RNA abundance and overall cell survival for each of the BARD1 variants. The score of 0 is considered to be functional for this assay, whereas the scores below zero 0 are non-functional.
Submission format
The prediction submission is a tab-delimited text file. Organizers provide a template file, which must be used for submission. A validation script is also provided; predictors must check the correctness of the format before submitting their predictions.
- Chromosome
- Position
- Reference allele
- Alternate allele
- Consequence (UTR variant, missense, synonymous, etc.)
- Variant using the HGVS protein nomenclature if available; e.g., p.Val695Leu, relative to protein sequence NP_000456.2 from RefSeq MANE select release v1.4.
- RA: RNA abundance score
- SDRA: for the abundance score; a positive number
- SS: Overall survival score
- SDSS: Standard deviation for the overall survival score; a positive number
- Comment: optional brief comment on the basis of the prediction in columns 2-4.
In the template file, cells in columns 7-11 are marked with a "*". Submit your predictions by replacing the "*" with your value. No empty cells are allowed in the submission. If you are not confident in a prediction for an individual, enter a suitably large standard deviation for the prediction. Optionally, enter brief comments indicating the basis of the predictions; otherwise, leave the "*" in these cells.
Please make sure you follow the submission guidelines strictly.
In addition, your submission must include a detailed description of the method used to make the predictions, similar to the style of the Methods section in a scientific article. This information will be submitted as a separate file.
File naming
CAGI allows submission of up to six models per team, of which model 1 is considered primary. You can upload predictions for each model multiple times; the last submission before deadline will be evaluated for each model. If you are submitting a single file with all predictions combined, please use the format below.
Use the following format for your submissions: <teamname>_model_(1|2|3|4|5|6).(tsv|txt)
To include a description of your method, use the following filename: <teamname>_desc.*
Example: if your team’s name is “bestincagi” and you are submitting predictions for your model number 3, your filename should be bestincagi_model_3.txt.
Download data
Variant data: available from the Synapse portal
Download submission template file: bard1submissiontemplate.tsv (provided on Synapse)
Download submission validation script: bard1validation.py (provided on Synapse)
Training data
No training data is provided. Participants may wish to use resources such as MaveDB, ClinVar, gnomAD, HGMD, UniProtKB, etc. to develop and calibrate their models.
Assessment
Predictions will be assessed by an independent assessor, Emidio Capriotti, University of Bologna. Evaluation metrics may include R-square, correlation, and rank correlation between predictions and experimental observations. Assessors may also compare the score distributions of predictions and observations.
Dataset provided by
Doug Fowler, Ivan Woo, and Lea Starita (University of Washington), leveraging their SGE platform previously applied to BRCA1.
References
Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev (2002) 12(1):86-91. PubMed
Findlay GM, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature (2018) 562(7726):217-222. PubMed
IGVF Consortium. Deciphering the impact of genomic variation on function. Nature (2024) 633(8028):47-57. PubMed
Irminger-Finger I, et al. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int J Biochem Cell Biol (2016) 72:1-17. PubMed
Tarsounas M, Sung P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol (2020) 21(5):284-299. PubMed
Revision history
4 June 2025: challenge preview posted
25 June 2025: edits in the challenge description, challenge open
30 July 2025: assessor information provided
15 September 2025: submission deadline extended from September 15 to September 30
30 September 2025: submission deadline extended from September 30 to October 15
19 October 2025: challenge closed on October 15